Diplaris, S.; Tsoumakas, G.; Mitkas, P.; Vlahavas, I. (2005). Protein Classification with Multiple Algorithms. In Proc. 10th Panhellenic Conference on Informatics, Volos, Greece, PCI05, 448--456.
genbase
mldr.datasets::get.mldr("genbase")
Summary
| Instances | 662 |
|---|---|
| Attributes | 1213 |
| Inputs | 1186 |
| Labels | 27 |
| Labelsets | 32 |
| Single labelsets | 10 |
| Max frequency | 170 |
| Cardinality | 1.2523 |
| Density | 0.0464 |
| Mean IR | 37.3146 |
| SCUMBLE | 0.0288 |
| TCS | 13.8399 |
Citation
@inproceedings{,
title = "Protein Classification with Multiple Algorithms",
author = "Diplaris, S. and Tsoumakas, G. and Mitkas, P. and Vlahavas, I.",
booktitle = "Proc. 10th Panhellenic Conference on Informatics, Volos, Greece, PCI05",
year = "2005",
pages = "448--456"
}Concurrence plot
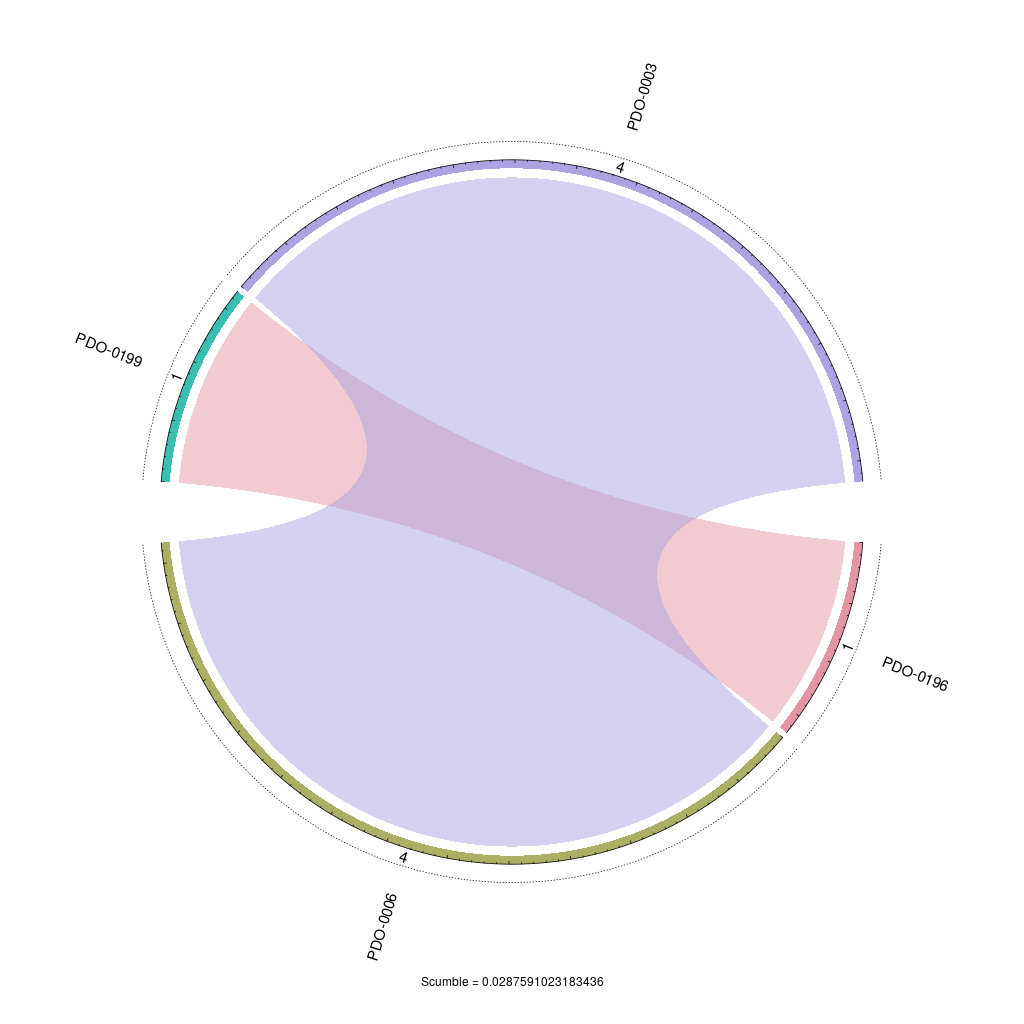
In this concurrence plot, sectors represent labels and links between them depict label co-occurrences. SCUMBLE is a measure designed to assess the concurrence among imbalanced labels.